Abstract

The molecular landscape of B cell precursor acute lymphoblastic leukemia (BCP-ALL) describes >20 disease subtypes defined by initiating genomic alterations and corresponding gene expression signatures. Some subtypes represent phenocopies with shared gene expression signatures but distinct or absent genomic drivers. The majority of BCP-ALL subtypes have been acknowledged as diagnostic entities by the currently revised classifications of hematologic malignancies (WHO-HAEM5 / International Consensus Classification). Thus, unified diagnostic approaches are needed to systematically identify subtypes in individual BCP-ALL cases in clinical routine. Gene expression signatures represent the shared functional equivalent of heterogeneous genomic driver alterations. Therefore, we established a gene expression-based classifier to identify BCP-ALL subtypes and related clinical and biological phenotypes.
We analyzed a total of n=3477 BCP-ALL gene expression profiles obtained from transcriptome sequencing of adult (n=1253) and pediatric patients (n=2224) including publicly available (n=2074; Gu Z et al. Nat. Genet. 2019; Schmidt B. et al. Blood Adv. 2022) and own data sets (n=1403) from adult and pediatric European study groups (GMALL, AIEOP-BFM, CELL). Raw read counts from 6 cohorts representing heterogeneous library preps (i.e., stranded/unstranded; mRNA/total RNA), sequencing platforms/depths, and gene count quantifications were included. All samples had been previously allocated to molecular BCP-ALL subtypes based on integration of independent genomic profiling, gene fusion calling and gene expression. For classification, data with defined subgroup allocation were randomly split into training (n=1831; 70%) and testing sets (n=785; 30%) and two hold-out studies (n=305).
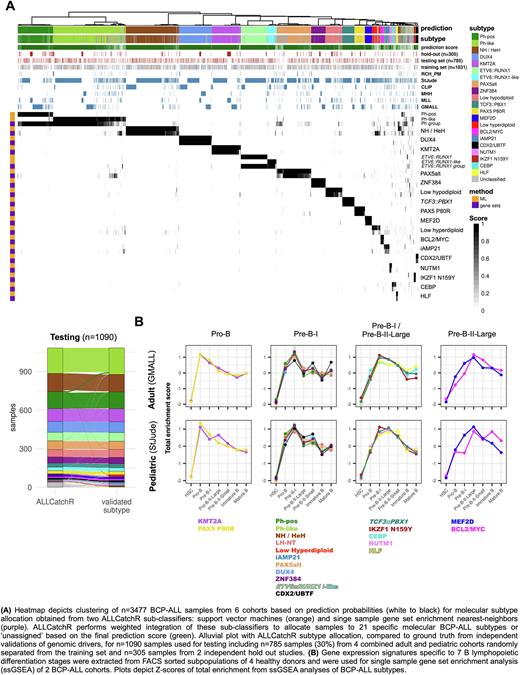
To define gene expression signatures of 21 BCP-ALL subtypes, we used training set samples and extracted specific gene sets employing different feature selection algorithms as well as ranking of genes in individual subtypes. Based on these definitions we established a BCP-ALL subtype classifier (ALLCatchR), which integrates machine learning models (support vector machines) and nearest-neighbors associations of single sample gene set enrichment analyses (Fig. A). ALLCatchR performed molecular subtype allocation for the testing set at high sensitivity (0.934±0.165) and specificity (0.997±0.005) on average across subtypes. A similar performance was achieved in the two hold-out cohorts despite completely independent data structures which have not been shown to the classifier (sensitivity: 0.946±0.097 / specificity: 0.996±0.007). Overall accuracies of 0.952 and 0.92 for testing and hold-out-data sets indicate that ALLCatchR reliably identifies molecular BCP-ALL subtypes in mixed adult / pediatric cohorts based on gene expression profiles alone (Fig. A, below). Among n=44/1090 (4%) samples of the combined hold-out/testing set with ambiguous subtype allocation (deemed 'unclassified'), ALLCatchR provides candidate subtype allocations (sensitivity: 0.671±0.393 / specificity: 0.978±0.036) for confirmation based on genomic driver alterations. In addition, we modified our classifier to predict baseline variables such as patient's sex (accuracy: 0.991), immunophenotype (accuracy: 0.935) or blast count (R=0.633). To define the corresponding B cell differentiation stage of origin, we compared BCP-ALL subtypes to differentiation stage-specific gene sets extracted from 7 FACS-sorted lymphopoietic subsets from healthy bone marrow donors (n=4). We observed defined BCP-ALL subtype-specific enrichment patterns which can be grouped along 4 major differentiation stages (Pro-B to Pre-B-II-Large, Fig. 1B) with a high degree of similarity in independent adult and pediatric cohorts. Thus, B cell developmental trajectories underlying BCP-ALL subtypes are dissected at high resolution by gene expression profiling and differentiation stages may favor the selection of specific leukemogenic drivers.
ALLCatchR is a gene expression classifier for 21 established BCP-ALL molecular subtypes, which performs at independently validated cohorts with an accuracy of >90% and allows imputation of clinical baseline variables as well as underlying B cell developmental trajectories. It provides a reliable basis for systematic diagnostic approaches in pediatric and adult BCP-ALL patients.
Disclosures
Walter:MLL Munich Leukemia Laboratory: Current Employment. Schrappe:Amgen: Research Funding; SigmaTau: Research Funding; Novartis: Honoraria, Research Funding; SHIRE: Research Funding; Servier: Honoraria, Research Funding; JazzPharma: Consultancy, Honoraria, Research Funding. Goekbuget:AbbVie: Other: Research funding (institution); AstraZeneca: Other: Invited talk for company-sponsored symposia (with honoraria); Novartis: Other: Research funding (institution); advisory board; Cellestia: Other: Advisory board; Erytech: Other: Advisory board; Gilead/Kite: Other: Research funding (institution); invited talk; advisory board; Incyte: Other: Research funding (institution); advisory board; Jazz Pharmaceuticals: Other: Research funding (institution); advisory board; MorphoSys: Other: Advisory board; Servier: Consultancy, Other: Research funding (institution); invited speaker; advisory board; Pfizer: Consultancy, Other: Research funding (institution); invited speaker; advisory board; Amgen: Consultancy, Other: Research funding (institution); invited speaker; advisory board; Clinigen: Other: Advisory board. Haferlach:MLL Munich Leukemia Laboratory: Current Employment, Other: Ownership. Bastian:BeiGene: Honoraria.
Author notes
 This icon denotes a clinically relevant abstract
This icon denotes a clinically relevant abstract
Asterisk with author names denotes non-ASH members.